Genetics, development and evolution of color and patterning
Our lab is working to understand the genetic and developmental basis of adaptive color variation in natural populations of mammals. We use a comprehensive approach, ranging from transcriptomics and genetic mapping to in vivo functional experiments, in order to uncover the genomic regions, and ultimately the genes and mutations, involved in phenotypic variation. These studies will allow us to address fundamental questions about evolutionary change: How many genes contribute to adaptive variation? Are the same genomic regions or different regions responsible for convergent phenotypes? Are these changes in regulatory or structural regions, and do they affect gene expression or protein function? What are the mechanisms by which these genes cause their effects?
Molecular basis of crypsis
We use different Peromyscus populations to uncover the genetic and developmental mechanisms underlying variation in cryptic coloration.
 - The Santa Rosa Island beach mouse (Peromyscus polionotus leucocephalus) and the oldfield mouse (P. p. subgriseus) from mainland Florida differ in pigmentation and patterning, each cryptically colored in their native habitat. Through QTL mapping approaches we have identified several genes that play a major role in explaining such differences. We are currently performing fine-scale mapping studies as well as functional experiments using lentiviral-based vectors in order to understand the mechanisms by which these molecules operate. (Photo credit: Clint Cook)
- The Santa Rosa Island beach mouse (Peromyscus polionotus leucocephalus) and the oldfield mouse (P. p. subgriseus) from mainland Florida differ in pigmentation and patterning, each cryptically colored in their native habitat. Through QTL mapping approaches we have identified several genes that play a major role in explaining such differences. We are currently performing fine-scale mapping studies as well as functional experiments using lentiviral-based vectors in order to understand the mechanisms by which these molecules operate. (Photo credit: Clint Cook)
 - Deer mice (P. maniculatus) inhabiting the light soils of the Nebraska Sandhills have unique banding patterns on their dorsal hairs which give them an overall golden color, relative to their closest relatives, and help them camouflage from visual predators. We use genome wide association studies in natural populations coupled to transgenic approaches in order to identify the genomic regions and specific mutations responsible for generating this adaptive change in coloration.
- Deer mice (P. maniculatus) inhabiting the light soils of the Nebraska Sandhills have unique banding patterns on their dorsal hairs which give them an overall golden color, relative to their closest relatives, and help them camouflage from visual predators. We use genome wide association studies in natural populations coupled to transgenic approaches in order to identify the genomic regions and specific mutations responsible for generating this adaptive change in coloration.
Molecular basis of pattern formation
Many mammals display a remarkable array of pigmentation patterns - from the stripes of a zebra to the spots of a leopard. Although we have a good understanding of the genetic and molecular regulation of pigment production, the mechanisms by which such patterns are established remain unknown. We are interested in understanding how changes during embryonic development can lead to different adult color patterns.
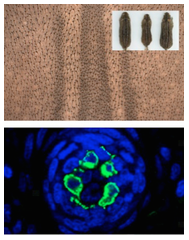 Using Peromyscus and other species with more complex color patterns, such as striped mice (Rhabdomys sp.) and chipmunks (Tamias), our lab employs a combination of transcriptomics, classic developmental approaches (in situ hybridization, qPCR, immunohistochemistry, etc…) and functional studies (genetics strains, ultrasound-guided in utero infection, etc…) to understand the cellular and molecular mechanisms underlying pattern formation. Here you can see the dorsal longitudinal stripes seen in striped mice during embryonic development and a Peromyscus DAPI-stained hair follicle with melanocytes stained in green. (Photo credit: Ricardo Mallarino and Marie Manceau, Harvard).
Using Peromyscus and other species with more complex color patterns, such as striped mice (Rhabdomys sp.) and chipmunks (Tamias), our lab employs a combination of transcriptomics, classic developmental approaches (in situ hybridization, qPCR, immunohistochemistry, etc…) and functional studies (genetics strains, ultrasound-guided in utero infection, etc…) to understand the cellular and molecular mechanisms underlying pattern formation. Here you can see the dorsal longitudinal stripes seen in striped mice during embryonic development and a Peromyscus DAPI-stained hair follicle with melanocytes stained in green. (Photo credit: Ricardo Mallarino and Marie Manceau, Harvard).
